Better Phylogenomic Models Resolve Long-Standing Mystery in Angiosperm Evolution
Published 29 October, 2024
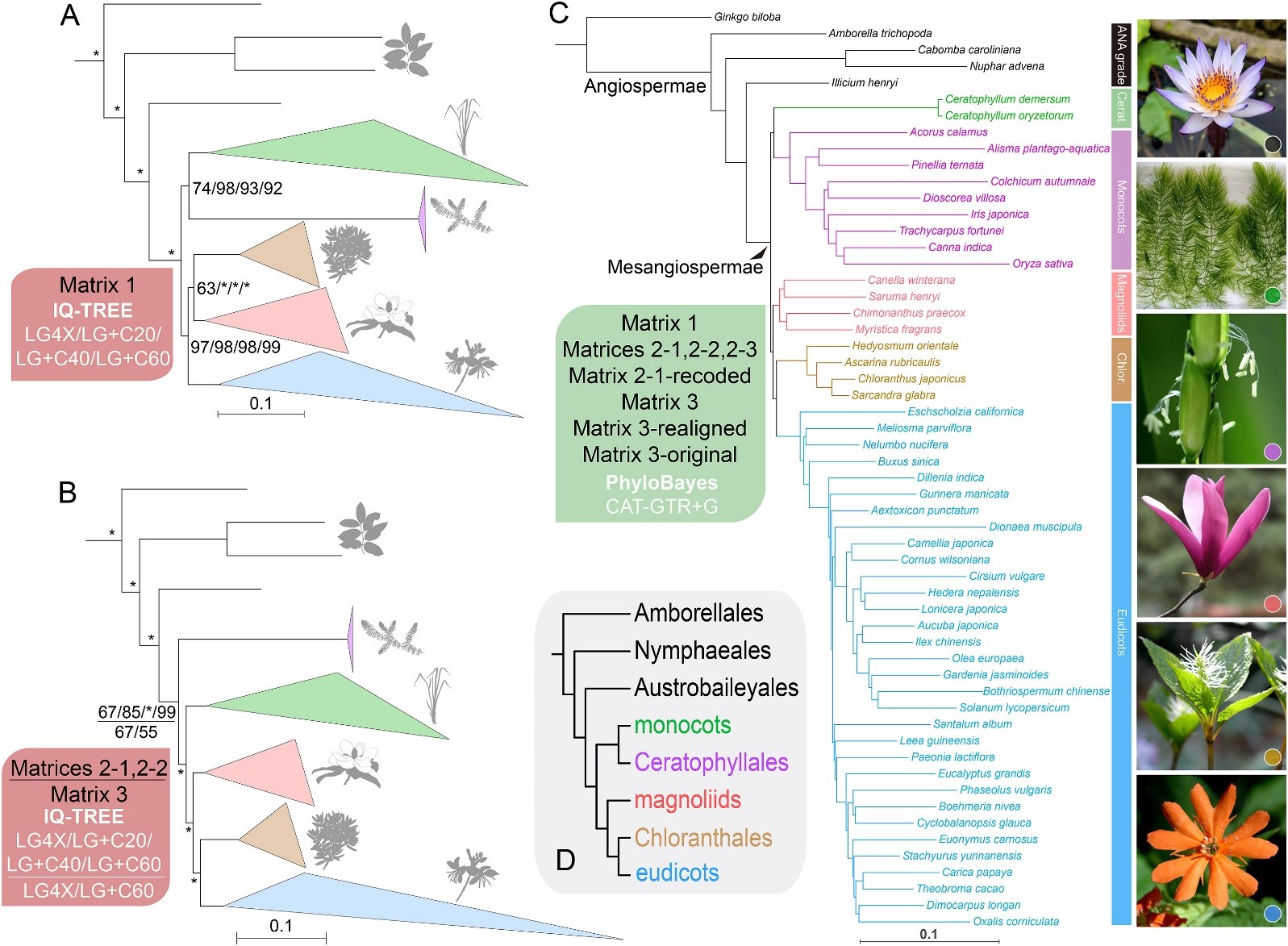
From towering trees and colorful garden flowers to crops like wheat, rice and fruits, angiosperms —or flowering plants — are everywhere, shaping ecosystems and feeding both humans and animals. Recent phylogenomic research, leveraging large-scale data from plastid, mitochondrial and nuclear genomes or transcriptomes, along with expanded taxon sampling, has provided insights into the evolutionary history of flowering plants. Despite these advances, the evolutionary relationships among the five major lineages of Mesangiospermae—comprising the vast majority of angiosperms—have remained unresolved due to the challenges posed by ancient, rapid radiations.
In a recent study, Chenyang Cai, a professor at the Nanjing Institute of Geology and Palaeontology, Chinese Academy of Sciences, and Yongli Wang from Jiangsu University, led a team to demonstrate that the relationships among five major mesangiosperm lineages can be resolved more accurately using genome-scale data with advanced models — even with plastid and mitochondrial genomes providing limited phylogenetic signals for deep evolutionary splits.
"Our analyses show that complex site-heterogeneous evolutionary models, which account for the functional constraints of proteins, outperform the traditional site-homogeneous or partition models commonly used in earlier studies," shares Cai.
Using advanced models within a Bayesian and maximum likelihood framework, the team identified a robust evolutionary tree. Ceratophyllales (commonly known as coontails) were revealed to be sister to monocots, and together they form a sister clade to magnoliids, Chloranthales and eudicots.
The study was published in the KeAi journal Plant Diversity.
“Our analysis of multiple molecular datasets highlights the importance of modeling molecular evolution to resolve rapid radiations in plants,” Cai explains. “The findings represent a significant breakthrough in understanding the evolutionary history of flowering plants, laying the groundwork for future ecological and evolutionary research.”
Contact author: Chenyang Cai, cycai@nigpas.ac.cn
Funder: The work has been supported by the National Natural Science Foundation of China (42222201, 42288201).
Conflict of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Article: Yongli Wang, et al., Modeling compositional heterogeneity resolves deep phylogeny of flowering plants, Plant Diversity, https://doi.org/10.1016/j.pld.2024.07.007